15. Elaborate how proteins' initiation, elongation, and termination happen in the ribosome with suitable diagrams.
15. Elaborate how proteins' initiation, elongation, and termination happen in the ribosome with suitable diagrams.
answer :
Introduction :
Ever wonder how antibiotics kill bacteria—for instance, when you have a sinus infection? Different antibiotics work in different ways, but some attack a very basic process in bacterial cells: they knock out the ability to make new proteins.
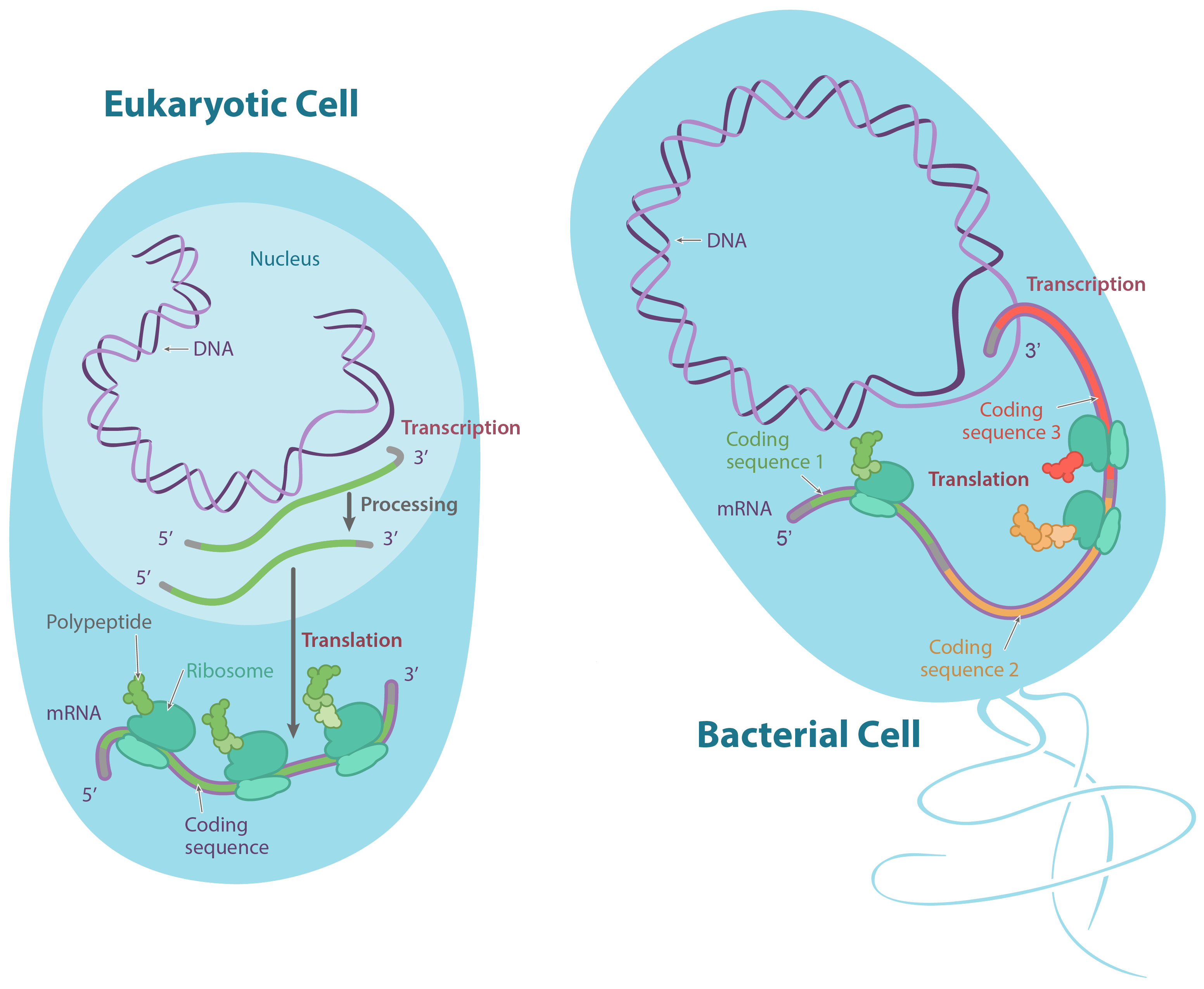
To use a little molecular biology vocab, these antibiotics block translation. In the process of translation, a cell reads information from a molecule called a messenger RNA (mRNA) and uses this information to build a protein. Translation is happening constantly in a normal bacterial cell, just like it is in most of the cells of your body, and it's key to keeping you (and your bacterial "visitors") alive.
When you take certain antibiotics (e.g., erythromycin), the antibiotic molecule will latch onto key translation molecules inside of bacterial cells and basically "stall" them. With no way to make proteins, the bacteria will stop functioning and, eventually, die. That's why infections clear up when they're treated with the antibiotic.start superscript, 1, comma, 2, end superscript
Cells need translation to stay alive, and understanding how it works (so we can shut it down with antibiotics) can save us from bacterial infections. Let's take a closer look at how translation happens, from the first step to the final product.
Translation: Beginning, middle, and end :
A book or movie has three basic parts: a beginning, middle, and end. Translation has pretty much the same three parts, but they have fancier names: initiation, elongation, and termination.
- Initiation ("beginning"): in this stage, the ribosome gets together with the mRNA and the first tRNA so translation can begin.
- Elongation ("middle"): in this stage, amino acids are brought to the ribosome by tRNAs and linked together to form a chain.
- Termination ("end"): in the last stage, the finished polypeptide is released to go and do its job in the cell.
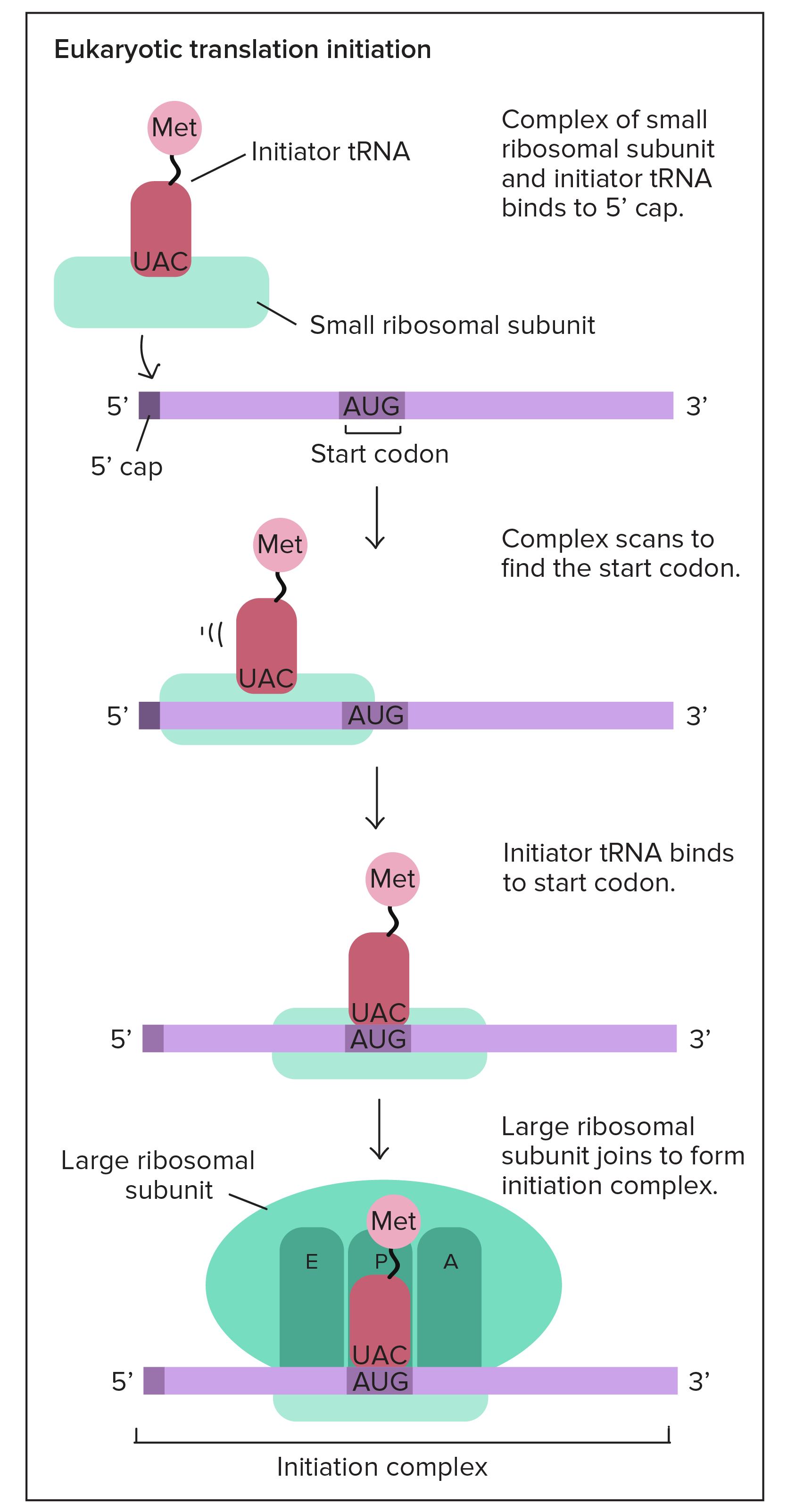
Initiation :
In order for translation to start, we need a few key ingredients. These include:
- A ribosome (which comes in two pieces, large and small)
- An mRNA with instructions for the protein we'll build
- An "initiator" tRNA carrying the first amino acid in the protein, which is almost always methionine (Met)
During initiation, these pieces must come together in just the right way. Together, they form the initiation complex, the molecular setup needed to start making a new protein.
Inside your cells (and the cells of other eukaryotes), translation initiation goes like this: first, the tRNA carrying methionine attaches to the small ribosomal subunit. Together, they bind to the 5' end of the mRNA by recognizing the 5' GTP cap (added during processing in the nucleus). Then, they "walk" along the mRNA in the 3' direction, stopping when they reach the start codon (often, but not always, the first AUG).start superscript, 6, end superscript
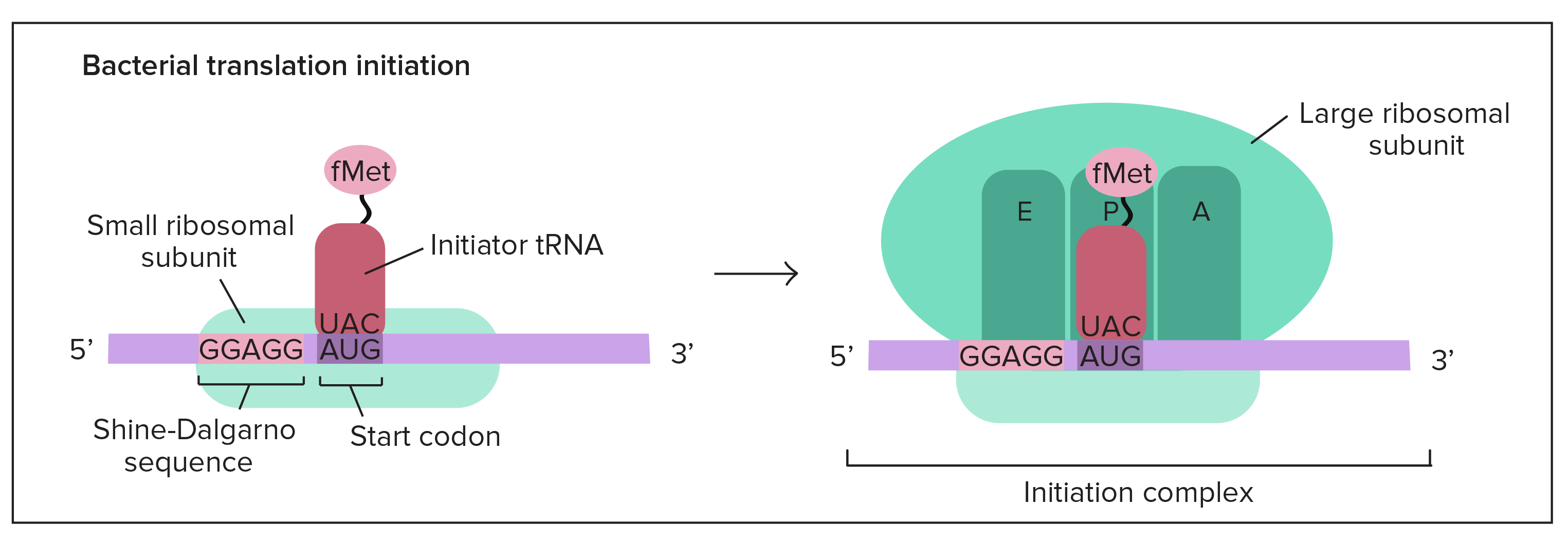
In bacteria, the situation is a little different. Here, the small ribosomal subunit doesn't start at the 5' end of the mRNA and travel toward the 3' end. Instead, it attaches directly to certain sequences in the mRNA. These Shine-Dalgarno sequences come just before start codons and "point them out" to the ribosome.
Why use Shine-Dalgarno sequences? Bacterial genes are often transcribed in groups (called operons), so one bacterial mRNA can contain the coding sequences for several genes. A Shine-Dalgarno sequence marks the start of each coding sequence, letting the ribosome find the right start codon for each gene.
Elongation :
I like to remember what happens in this "middle" stage of translation by its handy name: elongation is when the polypeptide chain gets longer.
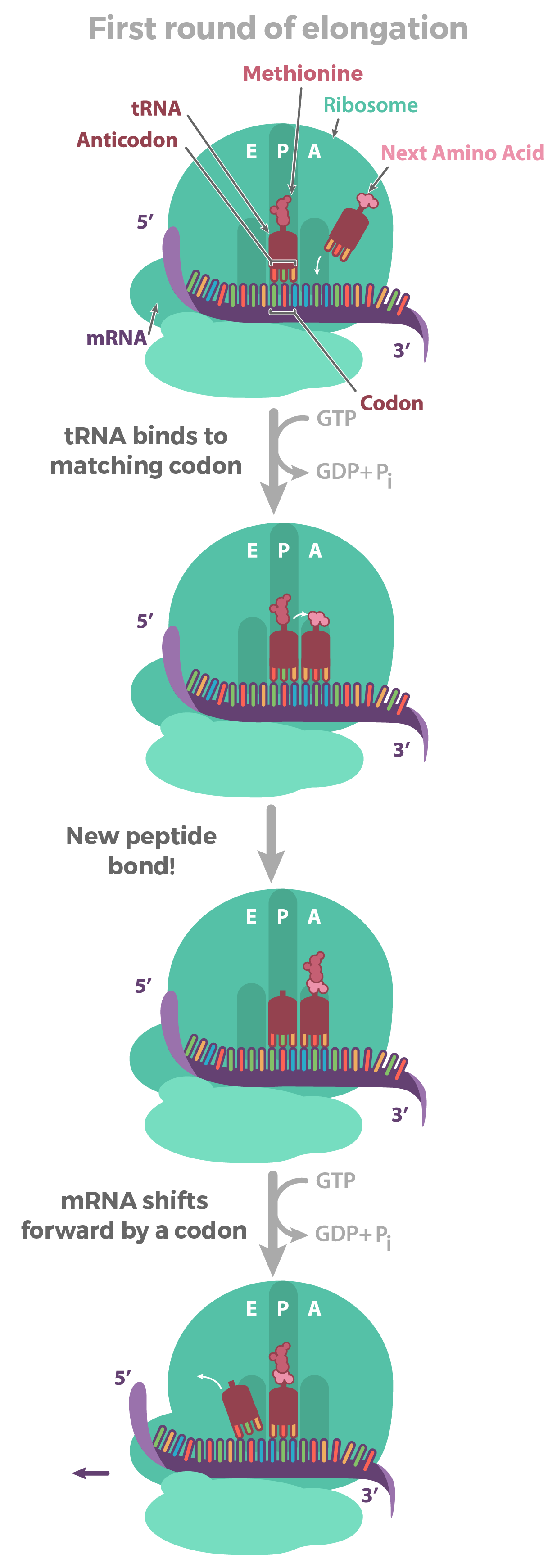
But how does the chain actually grow? To find out, let's take a look at the first round of elongation—after the initiation complex has formed, but before any amino acids have been linked to make a chain.
Our first, methionine-carrying tRNA starts out in the middle slot of the ribosome, called the P site. Next to it, a fresh codon is exposed in another slot, called the A site. The A site will be the "landing site" for the next tRNA, one whose anticodon is a perfect (complementary) match for the exposed codon.
Once the matching tRNA has landed in the A site, it's time for the action: that is, the formation of the peptide bond that connects one amino acid to another. This step transfers the methionine from the first tRNA onto the amino acid of the second tRNA in the A site.
Not bad—we now have two amino acids, a (very tiny) polypeptide! The methionine forms the N-terminus of the polypeptide, and the other amino acid is the C-terminus.
But...odds are we may want a longer polypeptide than two amino acids. How does the chain continue to grow? Once the peptide bond is formed, the mRNA is pulled onward through the ribosome by exactly one codon. This shift allows the first, empty tRNA to drift out via the E ("exit") site. It also exposes a new codon in the A site, so the whole cycle can repeat.
And repeat it does...from a few times up to a mind-boggling 33, comma000 times! The protein titin, which is found in your muscles and is the longest known polypeptide, can have up to 33, comma000 amino acidsstart superscript, 8, comma, 9, end superscript.
Termination :
Polypeptides, like all good things, must eventually come to an end. Translation ends in a process called termination. Termination happens when a stop codon in the mRNA (UAA, UAG, or UGA) enters the A site.
Stop codons are recognized by proteins called release factors, which fit neatly into the P site (though they aren't tRNAs). Release factors mess with the enzyme that normally forms peptide bonds: they make it add a water molecule to the last amino acid of the chain. This reaction separates the chain from the tRNA, and the newly made protein is released.
What next? Luckily, translation "equipment" is very reusable. After the small and large ribosomal subunits separate from the mRNA and from each other, each element can (and usually quickly does) take part in another round of translation.
conclusion :
Our polypeptide now has all its amino acids—does that mean it's ready to do its job in the cell?
Not necessarily. Polypeptides often need some "edits." During and after translation, amino acids may be chemically altered or removed. The new polypeptide will also fold into a distinct 3D structure, and may join with other polypeptides to make a multi-part protein.
Many proteins are good at folding on their own, but some need helpers ("chaperones") to keep them from sticking together incorrectly during the complex process of folding.
Some proteins also contain special amino acid sequences that direct them to certain parts of the cell. These sequences, often found close to the N- or C-terminus, can be thought of as the protein’s “train ticket” to its final destination.
Comments
Post a Comment