4. Elaborate in detail on leading and lagging strands in the DNA replication process and the essential components for DNA replication to occur?
4. Elaborate in detail on leading and lagging strands in the DNA replication process and the essential components for DNA replication to occur?
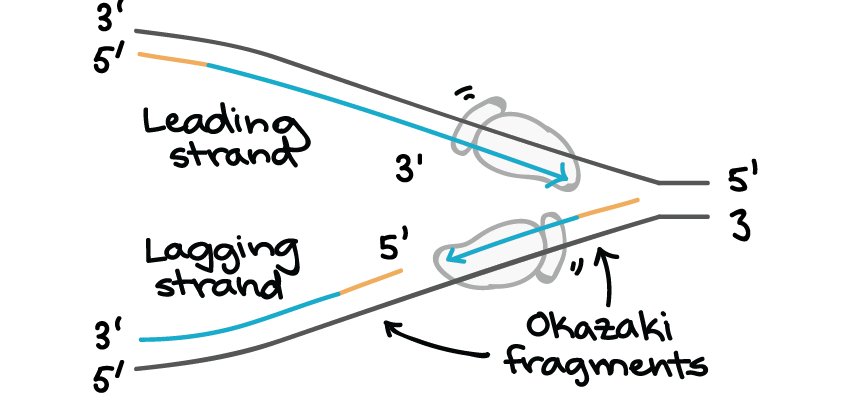
DNA replication is the first step of the central dogma where the DNA strands are replicated to make copies. During the process of replication the double stranded DNA is separated from each other by the help of enzymes like topoisomerases and helicases. The separated DNA strands form a replication fork, where both the DNA strands get replicated forming a lagging and leading strand.
The major difference between a lagging and leading strand is that the lagging strand replicates discontinuously forming short fragments, whereas the leading strand replicates continuously. Find out more such differences between a lagging and leading strand, in the table below.
leading and lagging strands in the DNA replication process :
Leading and lagging strands
Lagging Strand | Leading Strand |
Description | |
The strand that opens in the 3’ to 5’ direction towards the replication fork is referred to as the lagging strand. | The strand that runs in the 5’ to 3’ direction in the replication fork is referred to as the leading strand. |
Replication | |
The strand is replicated discontinuously. | The strand is replicated continuously. |
Fragments | |
Short stretches called okazaki fragments are formed during replication. | No short fragments are formed. |
Primer Requirements | |
Each fragment requires its own set of primers. | It requires only one primer. |
Ligase Requirement | |
It requires DNA ligase enzymes for the joining of short okazaki fragments. | It does not require DNA ligase. |
Direction of Growth | |
It grows away from the replication fork. | It grows in the direction of the replication fork. |
Speed | |
The synthesis of new strands is slow. | The synthesis of new strands is fast. |
The essential components for DNA replication to occur :
There are four basic components required to initiate and propagate DNA synthesis. They are: substrates, template, primer and enzymes.
Substrates
Four deoxyribonucleotide triphosphates (dNTP's) are required for DNA synthesis (note the only difference between deoxyribonucleotides and ribonucleotides is the absence of an OH group at position 2' on the ribose ring). These are dATP, dGTP, dTTP and dCTP. The high energy phosphate bond between the a and b phosphates is cleaved and the deoxynucleotide monophosphate is incorporated into the new DNA strand.
Ribonucleoside triphosphates (NTP's) are also required to initiate and sustain DNA synthesis. NTP's are used in the synthesis of RNA primers and ATP is used as an energy source for some of the enzymes needed to initiate and sustain DNA synthesis at the replication fork.
Template
The nucleotide that is to be incorporated into the growing DNA chain is selected by base pairing with the template strand of the DNA. The template is the DNA strand that is copied into a complementary strand of DNA.
Primer
The enzyme that synthesizes DNA, DNA polymerase, can only add nucleotides to an already existing strand or primer of DNA or RNA that is base paired with the template.
Enzymes
An enzyme, DNA polymerase, is required for the covalent joining of the incoming nucleotide to the primer. To actually initiate and sustain DNA replication requires many other proteins and enzymes which assemble into a large complex called a replisome. It is thought that the DNA is spooled through the replisome and replicated as it passes through.
conclusion :
DNA replication is a highly regulated molecular process where a single molecule of DNA is duplicated to result in two identical DNA molecules. As a semiconservative process, the double helix is broken down into two strands, where each strand serves as the template for the newly synthesized strand by matching complementary bases. Because DNA polymerase III can only synthesize the new strands from 5′-3′, this results in a leading strand that is continuously synthesized and a lagging strand that requires the use of Okazaki fragments. Meanwhile, because eukaryotes have linear DNA, telomeres are needed to ensure genetic information is not lost during replication. Because DNA is critical to life, research continues to better understand and treat diseases caused by mutations and damages in an individual’s DNA.
Comments
Post a Comment